
Load image features into adata.obsm and compute Leiden clustering. Measure CellProfiler’s Granularity features within segments for each crop: Add MeasureGranularity module and define parameters.
Convert image crops to gray images: Add ColorToGray module and define parameters. Crops and segmentations files are aligned automatically. CP-Pipeline NamesAndTypes: Declare to load crops as color images and segmentations as objects.

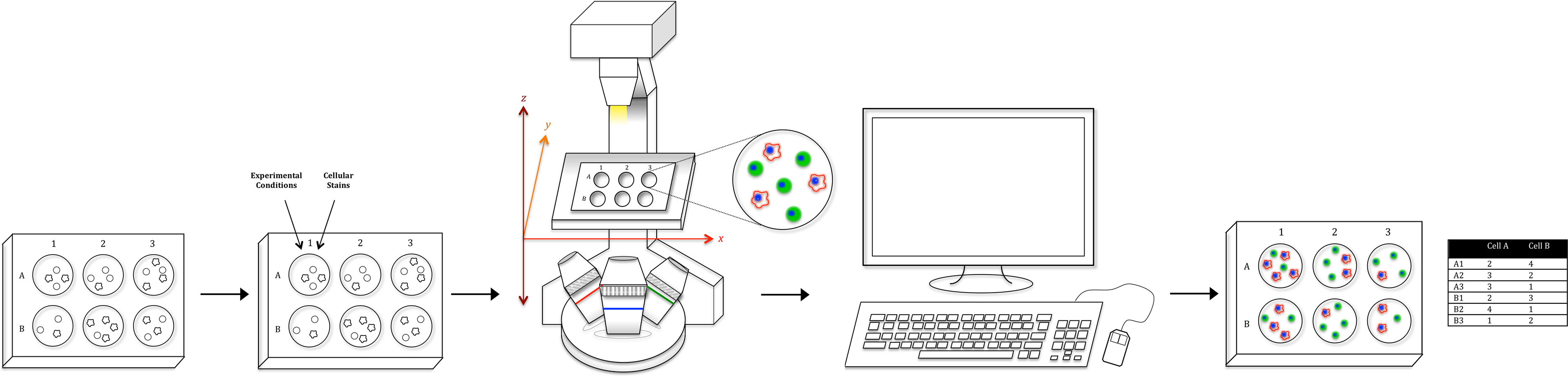
CP-Pipeline Images: Drag and Drop the folder imgs_dir into CellProfiler. tif' ) CellProfiler Pipeline: Calculate Image Features 1. obs_names, return_obs = True, as_array = False ): Image. The results of a pipeline execution is stored back in BisQue as table and image resources linked to the module execution. generate_spot_crops ( adata, obs_names = adata. Import packages & data įor crop, obs in img.
#Cellprofiler output during processing how to
For information on how to use the cellprofiler-core package for Python integration, the following Since this is not yet publicly well documented, we’ll restrict this tutorial to the laborious way of saving intermediate files to bridge Squidpy and CellProfiler.
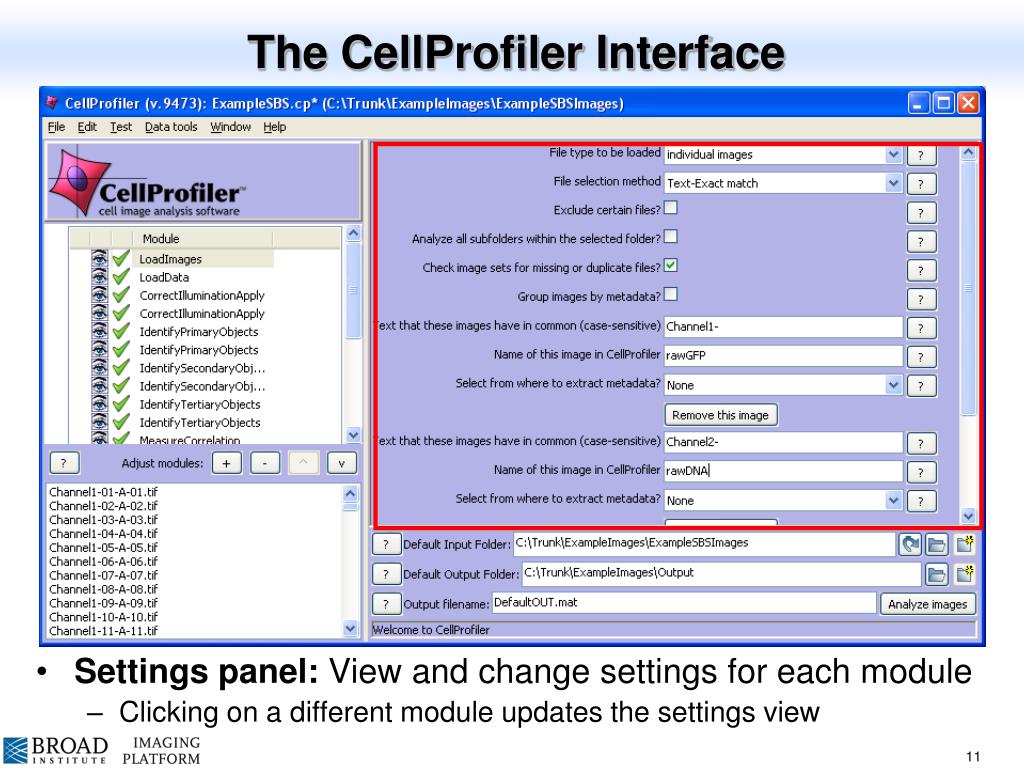
Note: In the future, CellProfiler functions will also be accessible via Python directly (according to the announcements of the CellProfiler team). Check the issues on CellProfiler Github in case of installation problems (can be tricky). First, download and install CellProfiler from the download page. In this tutorial, we show how to use Squidpy with functions from CellProfiler pipelines for image processing and feature extraction.Ĭalculate CellProfiler’s granularity features for image crops of Visium spots.Ĭompute clustering on the image features in Squidpy.ĬellProfiler is typically used via its GUI interface to build image processing pipelines.


 0 kommentar(er)
0 kommentar(er)
